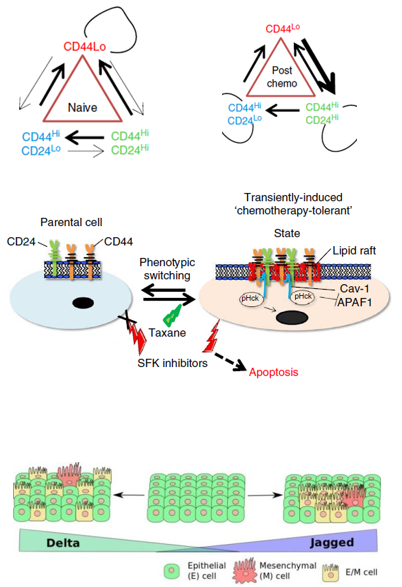
Project #1: Non-Darwinian dynamics affecting tumor immunity and drug resistance.
The tumor microenvironment is composed of immune cells, stromal cells and cancer cells, which can each influence one another to respond or resist to cancer treatment. While inherently drug resistant populations of cancer cells (sometimes referred to as cancer stem cells) have been classically ‘pin-pointed’ as the drivers of therapy failure and tumor re-growth, we recently identified that non-cancer stem cells (non-CSC) are predisposed to acquire resistance. We have now begun to unravel how immune cells are also affected by this non-mutational behavior to influence drug response, resistance and tumor immunity.
For example:
- How are immune cells influenced by anticancer therapies and cell-cell interactions?
- What are the genetic features of non-CSC that enable quick adaptation to a drug tolerant phenotype?
- How can we employ mathematical modeling to study population dynamics in the context of non-Darwinian drug resistance, and what tools to we need to validate these predictions experimentally?
- How does spatial heterogeneity contribute to phenotypic dynamics under drug pressure? And how does the stroma affect these transitions? Can they be targeted pharmacologically?
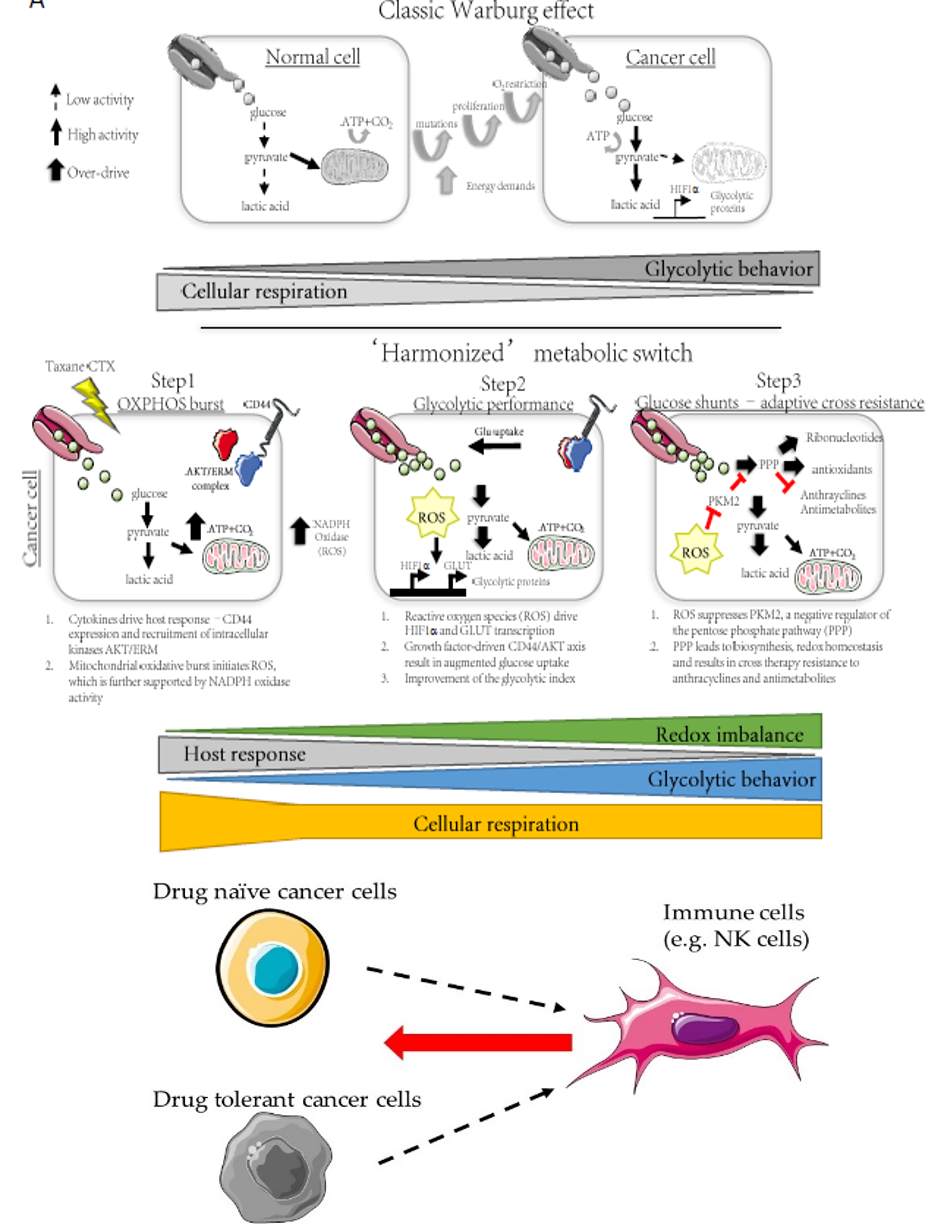
Project #2: Interrogating the role of metabolism in acquired drug resistance.
It is increasingly clear that cell metabolism can drive inherent mechanisms of drug resistance. However, it remains to be fully understood how dynamic cell states, particularly under drug pressure, can result in a rewiring of the metabolic phenotype of surviving cancer cells. Moreover, can this metabolic ‘shift’ confer acquired resistance to drug combinations? We recently discovered the some cancer cells will switch their phenotype under drug pressure to resemble a hybrid epithelial-mesenchymal phenotype, defined as a drug tolerant cell (DTC).
We are now seeking to address the following questions:
- How does the metabolic state of drug naïve cancer cells compare to the metabolic state of drug tolerant cancer cells (DTC)?
- Do different drugs induce different metabolic states in the persisting population of DTC? How do immunotherapies influence these dynamics?
- How does a cell’s metabolic phenotype influence the tumor-immune contexture? What tools can we employ to interrogate this heterogeneity?
- What new druggable targets will emerge that can be suppressed or enhanced to overcome drug resistance?
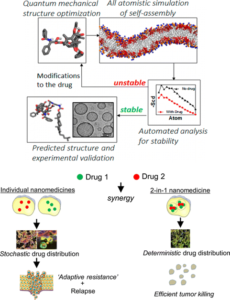
Project #3: Engineering novel therapies to combat drug resistance using nanotechnology.
While the Drug Resistance Group puts a heavy emphasis on the biological underpinning of therapy failure, we have a unique focus to develop novel therapies that can be translated immediately to the clinic. In collaboration with Dr. Shiladit Sengupta and Dr. Ashish Kulkarni, we recently demonstrated that supramolecular assembly of lipid nanoparticles can be informed through a computational approach. Using this strategy we addressed drug resistance by integrating a combination of drugs together into a single nanovehicle. This technology enables us to explore engineering approaches in greater depth, and to address mechanisms of resistance using next-generation technologies.
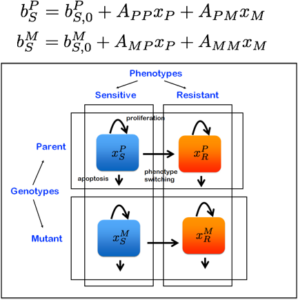
Project #4: Modeling phenotypic heterogeneity and cell fitness. In Collaboration with Dr. Martin Nowak, Ph.D.
The tumor ecology is a cohesive mixture of cancer cells, normal cells, and a microenvironment comprised of growth factors cytokines and chemokines. This heterogeneous ecosystem is essential to therapy response, and mechanisms of resistance are directly related to the diverse makeup of the tumor.
In this project we have integrated mathematical modeling and game theory, next-generation sequencing and other bioengineering tools to study cell fitness at the single cell level.
Are there novel mechanisms of resistance that are conferred under drug pressure, which can’t be known through conventional experimental approaches? What is the role of each cell type in this ‘game’?
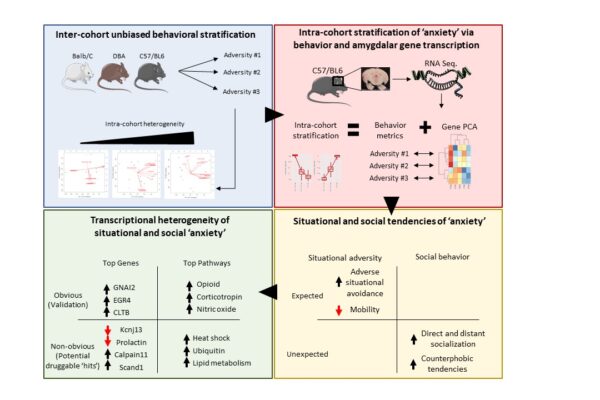
Project #5: Math, Mice and Anxiety — Leveraging in-vitro and in-vivo models to study anxiolytic response and resistance mechanisms in collaboration with Dr. Stephen Moss, Ph.D.
In collaboration with Dr. Stephen Moss (Tufts Medical School), The DRG and Drs. Ilana Braun (Brigham/DFCI Psychiatry) and Luke Lee (Harvard/Engineering) are investigating novel brain organoid models and in-vivo experiments to develop better anxiolytics. The team deploys computationally inspired models that leverage RNA sequencing, behavior experiments and bioinformatics to elucidate the transcriptomic signatures underpinning drug response. We use a unique class of phyto-chemicals (plant derived cannabinoids and terpenes) to uncover novel treatment strategies for psychiatric illnesses.